Tutorial -- Xray Absorption Spectroscopy with TDDFT
Performing a parallel relativistic calculation of excitation energies and Xray electronic absorption spectra (XAS) with conventional TDDFT is a simple two-step process
respect --scf --inp=my-input-file --nt=4 --scratch=/path/to/scratch/directory respect --tddft --inp=my-input-file --nt=4 --scratch=/path/to/scratch/directory
The first call of respect with the argument --scf performs the self-consistent field (SCF) optimization of ground-state molecular orbitals, which is a necessary prerequisite for the following property calculation. The second call of respect with the argument --tddft actually performs the TDDFT simulation.
The other arguments mandatory to respect are
--inp
specifies a name of the input file;--nt
specifies the number of physical cores required for parallel execution;--scratch
specifies a path to the scratch directory.
The input file my-input-file.inp should contain the input block scf: with some SCF-specific keywords, and the input block tddft: with some TDDFT-specific keywords. A comprehensive list of all keywords can be found for SCF here and for TDDFT here. A simple example of my-input-file.inp looks like
#SCF section
#DFT calculation of H2S molecule, employing the relativistic 2c
#amfX2C Hamiltonian, PBE0 functional and uncontracted cc-pvdz basis
scf:
geometry:
S 0.000000 0.000000 0.000000
H 0.000000 0.000000 1.336000
H 1.335103 0.000000 -0.048956
method: ks-amfX2C/pbe0
basis:
H: ucc-pvdz
S: ucc-pvdz
charge: 0
multiplicity: 1
maxiterations: 30
nc-model: gauss
convergence: 1.0e-7
#TDDFT section
tddft:
num-eigenvalues: 48
print-level: debug
convergence: 1.0e-4
#selection of a window of MOs within which excitations can take place
mowindow:
type: cvs #core-valence separation
occupied: 5-10 #occupied MOs
virtual: 19-44 #virtual MOs
Final output data from the TDDFT calculation are saved in file my-input-file.out_tddft. Of relevance for XAS are oscillator strengths evaluated for the lowest 48 excitation energies involving the selected occupied-virtual MOs. An example of the output block for the first excitation energy reads
================
Excited state: 1
================
::
Excitation energy [units]
-------------------------
5.8414 [ a.u. ]
158.9531 [ eV ]
7.8000 [ nm ]
15336.6485 [ kJ/mol-1 ]
1282043.5931 [ cm-1 ]
Oscillator strength [dimensionless]
-----------------------------------
x 0.0000000000
y 0.0000003646
z 0.0000000000
Trace 0.0000003646
These data can be used to plot the final XAS spectrum. ReSpect offers a simple python script EAS_from_TDDFT.py to do that
/path/to/ReSpect/util/EAS_from_TDDFT.py --files=my-input-file --damping=0.01 --window="158-170" --stick --plot
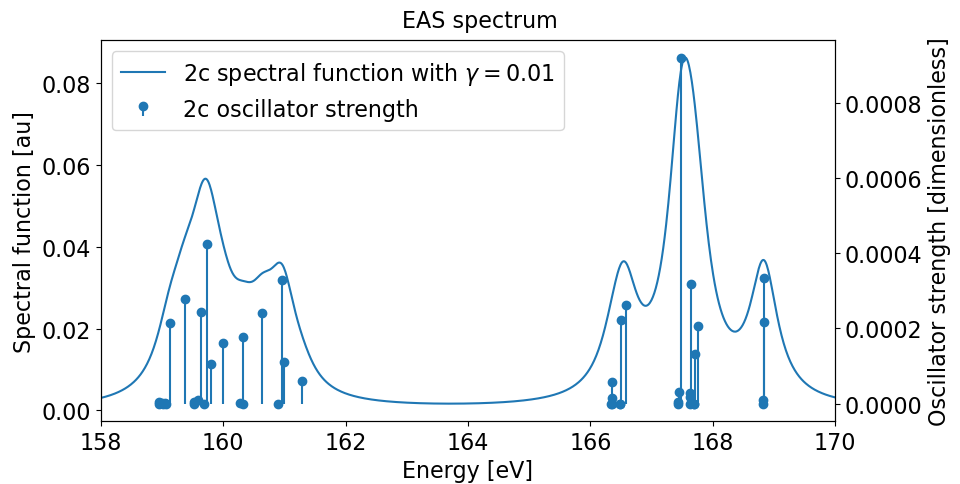
In addition, ReSpect also provides a flexible restart scheme for TDDFT calculations. Possible reasons for the need to restart TDDFT include (but are not restricted to)
previous calculations did not finish successfully
better convergence threshold is desired
increasing the number of excitation energies
increasing the complexity of the TDDFT method (restart from Tamm--Dancoff approximation)
Following the example above, all data required for the TDDFT restart are stored in file my-input-file.50. In analogy with the SCF restart scheme, one needs to create a soft link between the old data file my-input-file.50 and a new data file, as well as to copy the old input file my-input-file.inp to a new input file
ln -sf my-input-file.50 my-restart-file.50 cp my-input-file.inp my-restart-file.inp
Now, one can modify the TDDFT input block in my-restart-file.inp -- for example, increase the number of excitation energies and tighten the convergence threshold
tddft:
num-eigenvalues : 62
convergence : 5.0e-5
and then restart the TDDFT calculation by running
respect --restart --tddft --inp=my-restart-file --nt=4 --scratch=/path/to/scratch/directory
Finally, there are several important aspects associated with the input syntax, namely
the input is case-insensitive
This means that the program does not distinguish between uppercase and lowercase letters.the input is insensitive to the number of blank lines and/or comment lines
All comments begin with the number sign (#), can start anywhere on a line and continue until the end of the line.the input is compliant with the dictionary syntax of the YAML markup language
This means that each input line is represented either by a singleblock:statement or by a simplekeyword:valuepair, such as
block1:
keyword1: value1
keyword2: value2
...
block2:
keyword3: value3
keyword4: value4
...
block3:
keyword5: value5
keyword6: value6
...
It is essential to remember that all members of one block: are lines beginning at the same indentation level. Whitespace indentation is used to denote the block structure; however, tab characters are never allowed as indentation. The only exception to the YAML-based input syntax is the block geometry: which utilizes a simple xyz format for the molecular geometry specification.
TIPS & TRICKS
Q: How to scale the speed of light in TDDFT calculations?
Set the cscale option in the SCF calculation. The scaling value is then automatically transferred to the TDDFT calculation.
Q: Is there a way to launch SCF and TDDFT calculations without the need to explicitly setup the scratch path by "--scratch=/path/to/scratch/directory"?
Yes, the argument "--scratch=/path/to/scratch/directory" can be saved to the file .respectrc in your home directory. If both the file and the command line argument exist, then ReSpect takes the scratch directory setting from the command line.
Q: How to recover crashed jobs?
Find the path to the scratch directory of your job (it is printed on the standard output (stdout)) and run the command
respect --recover-job=/path/to/scratch/directory/of/your/job
The script will create file respect_checkpoint_file.50, which contains all necessary restart information. Follow the restart procedure described in the above tutorial.
Latest Publications
X2C Hamiltonian Models in ReSpect: Bridging Accuracy and Efficiency
Light-induced electronic chirality in achiral molecules
Useful Links
Our Contacts
Department of Chemistry
UiT The Arctic University of Norway
Tromsø, NO-9037 Norway
Email: info@respectprogram.eu